Global minimization, 1d, assignment 4 a), parallel (parfor, spmd)
12.3.2018 apiola@triton:2018kevat/Heikki/Lecture4/Globalminsolve2.m
Continuation to Globalminsolve1.m
Contents
- Function to minimize
- Task:
- Define objective function:
- Split into several parts:
- Let's make use of all 24 workers on Triton, take wider range.
- fminbnd, only bounds are needed, no starting points.
- Observations:
- Parallel computing with spmd
- spmd
- Compare with using just Matlab's vector function min.
- Comments, conclusions, visions
Function to minimize
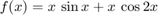
Find global minimum (and local minima) on [-2,14]. Split the interval into pieces and use fminbnd on each piece.
Task:
Enlarge interval to [-2,14] (a) Change for to parfor, run on pc and Triton, do tic - toc- timing. (b) Change to spmd, on Triton you can take more labs than 6. (c) Find max-points as well. Bounds: lb = -2; ub = 14;
Here we will do some more and some less (let's leave the max-part).
clear close all format compact
Define objective function:
f = @(x) x.*sin(x) + x.*cos(2.*x)
f =
function_handle with value:
@(x)x.*sin(x)+x.*cos(2.*x)
Split into several parts:
%{ lb=[-2 0 1 3 6 8 10 12]; % Lower bounds ub=[lb(2:end) 14]; % Upper bounds % x0=0.5*(lb+ub); % Starting points for solver that requires them. N=length(lb); % Number of subintervals, call them "labs". %}
Let's make use of all 24 workers on Triton, take wider range.
Remove(d) comments for this 2^nd run
Low=-35;Up=35;N=24; lb=linspace(Low,Up,N+1); ub=lb(2:end); lb=lb(1:end-1);
fminbnd, only bounds are needed, no starting points.
Basic use: [xmin,ymin]=fminbnd(f,lb,ub);
xmin=zeros(N,1); ymin=xmin; % parpool; % Remove comment if pool not open. tic % Move commments to parfor and back % Comment away plot-commands with parfor and when comparing timings. %for k=1:10 % Take 10 runs to have average timing. %for i=1:N parfor i=1:N [xmin(i),ymin(i)] = fminbnd(f,lb(i),ub(i)); % No graphics now. %{ subplot(ceil(N/2),2,i) fplot(f,[lb(i) ub(i)]); grid on hold on plot(xmin(i),ymin(i),'ro') % Plot "labwise" minimum point (red circle) hold off %} end toc %{ N = 24 Elapsed time is 1.479764 seconds. Elapsed time is 0.154422 seconds. %}
Elapsed time is 0.866446 seconds.
Observations:
- 2^nd (or 3^rd) run is much faster then the 1^st, not to speak about the pool opening run.
- There is little difference with N=8 to N=24 (parfor shows its strength)
- There is little (if any) difference to for, too much overhead compared to intensive computation. Need examples of "heavier" funs.
Tfor(k)=toc; Tparfor(k)=toc; end meanTfor=mean(Tfor) meanTfor = 0.2513 meanTparfor=mean(Tparfor) % First call very slow, setup of pool with workers meanTparfor = 0.5459 % Still 2 x slower, gosh!
minptspar=[xmin ymin];
close all
figure fplot(f,[Low,Up]) hold on plot(xmin,ymin,'*r');grid on;shg %
Parallel computing with spmd
spmd
My very first spmd-block
spmd Lind=labindex; end Lindcell=Lind(1:5) % First five cells Lindvect=[Lind{:}] % % To use spmd with fminbnd, we need a form whose argument list is a % 2-vector instead of two scalars. So, here: fminbndI=@(f,Interval) fminbnd(f,Interval(1),Interval(2)) % (On the 2-variable optimization we need the converse.) %
Lindcell =
1×5 cell array
{[1]} {[2]} {[3]} {[4]} {[5]}
Lindvect =
Columns 1 through 13
1 2 3 4 5 6 7 8 9 10 11 12 13
Columns 14 through 24
14 15 16 17 18 19 20 21 22 23 24
fminbndI =
function_handle with value:
@(f,Interval)fminbnd(f,Interval(1),Interval(2))
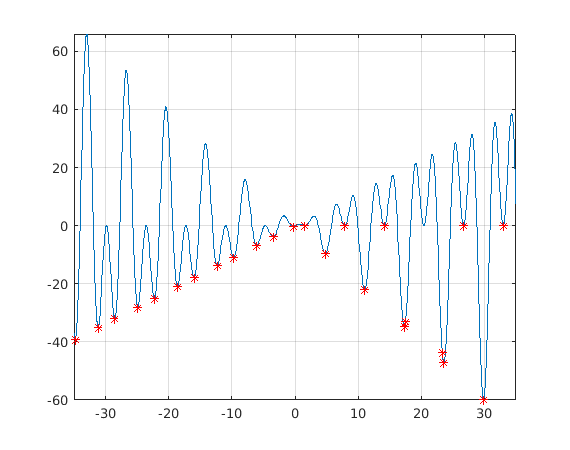
tic spmd intervals=[lb' ub']; Int=intervals(labindex,:); [xminspmd,yminspmd]=fminbndI(f,Int); end Tspmd=toc % 0.24... Slightly better than for, twice faster than parfor. % % And NOTE the ELEGANCE! minpts_spmd=[xminspmd{:};yminspmd{:}]; % From workers to client. %
Tspmd =
0.4513
figure fplot(f,[-35,35]);grid on hold on plot(minpts_spmd(1,:),minpts_spmd(2,:),'.r','MarkerSize',10) shg
Compare with using just Matlab's vector function min.
%figure tic for i=1:N % parfor i=1:N xx=linspace(lb(i),ub(i)); yy=f(xx); ymin=min(yy); Yes=(yy==ymin); % Better test: abs(yy-ymin)< tol, see later xmins{i}=xx(Yes); ymins{i}=yy(Yes); %{ subplot(4,2,i) fplot(f,[lb(i) ub(i)]); grid on hold on plot(xmins{i},ymins{i},'ro') % Plot "labwise" minimum point (red circle) %hold off %} end display(N) Tfor=toc % 0.005 %Tparfor=toc % 0.14... minptsvecop=[xmins{:};ymins{:}];
N =
24
Tfor =
0.0206
Comments, conclusions, visions
Simple vector operation was about as fast (or even faster) and all results were correct, contrary to fmincon, which got the endpoint values on "labs" 6 and 8 wrong (N=8).
If you want to increase accuracy, you will do better by adjusting the tolerances with optimset, and running fmincon with tight intervals (obtained with logical indexing as above).
Vector operations are so fast that using this technique there's little point in splitting them up into "labs" in case only global minimum is sought unless the data is very big or some parts of the domain require much more dense discterization than some others. (Recall adaptivity with ODE-solvers).
Here we have an example of several local minima, which requires the kind of splitting we have done
Several varibles and parameters increse the benefits of parallelization.