Global minimization, 1d, assignment 4 a) Globalminsolve1.m
12.3.2018 apiola@triton:2018kevat/Heikki/Lecture4/
Contents
Function to minimize
Find global minimum (and local minima) on [-2,14]. Split the interval into pieces and use fminbnd on each piece.
Task:
Enlarge interval to [-2,14]
- Change for to parfor, run on pc and Triton, do tic - toc- timing.
- Change to spmd, on Triton you can take more labs than 6.
- Find max-points as well.
Bounds: lb = -2; ub = 14;
Here we will do some more and some less (let's leave the max-part).
clear close all format compact
Define objective function:
f = @(x) x.*sin(x) + x.*cos(2.*x)
f =
function_handle with value:
@(x)x.*sin(x)+x.*cos(2.*x)
Split into several parts:
lb=[-2 0 1 3 6 8 10 12]; % Lower bounds ub=[lb(2:end) 14]; % Upper bounds % x0=0.5*(lb+ub); % Starting points for solver that requires them. N=length(lb); % Number of subintervals, call them "labs". %
fminbnd, only bounds are needed.
Basic use: [xmin,ymin]=fminbnd(f,lb,ub);
xmin=zeros(N,1); ymin=xmin; % Preallocation % parpool; % Remove comment if pool not open. tic % Move commments to parfor and back % Comment away plot-commands with parfor and when comparing timings. %for k=1:10 % Take 10 runs to have average timing. for i=1:N %parfor i=1:N [xmin(i),ymin(i)] = fminbnd(f,lb(i),ub(i)); % %{ subplot(ceil(N/2),2,i) fplot(f,[lb(i) ub(i)]); grid on hold on plot(xmin(i),ymin(i),'ro') % Plot "labwise" minimum point (red circle) hold off % %} end toc
Elapsed time is 0.432004 seconds.
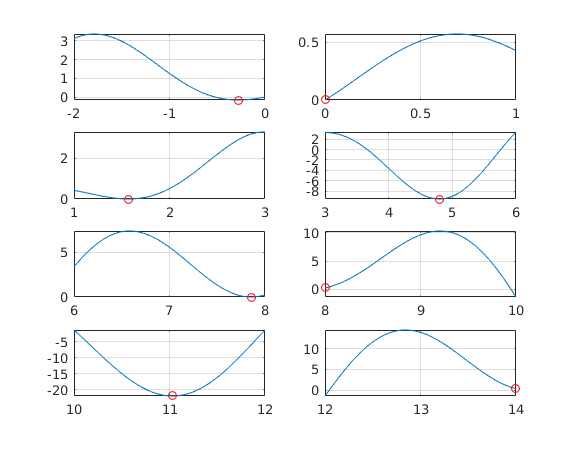
Notice: "Labs" 6 and 8 have the endpointminima wrong, but for the whole the bdary points are often artificial.
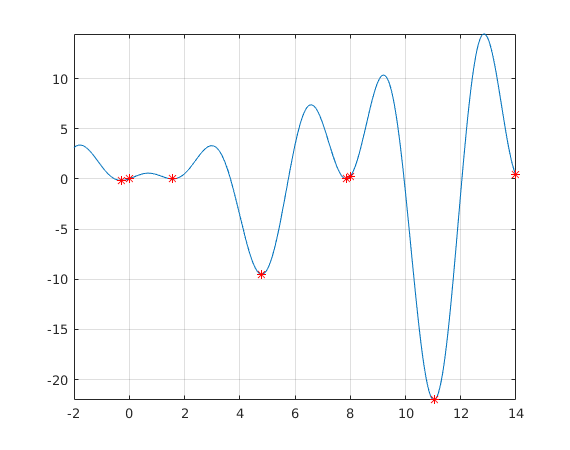
%{ N = 24 Elapsed time is 1.479764 seconds. Elapsed time is 0.154422 seconds. %} minpts=[xmin ymin] %
minpts =
-0.2809 -0.1599
0.0001 0.0001
1.5708 0.0000
4.7954 -9.5084
7.8540 0.0000
8.0000 0.2538
11.0318 -22.0274
14.0000 0.3923
Observations:
- 2^nd (or 3^rd) run is much faster then the 1^st, not to speak about the pool opening run.
- There is little difference with N=8 to N=24 (parfor shows its strength)
- There is little (if any) difference to for, too much overhead compared to intensive computation. Need examples of "heavier" funs and/or bigger data.
%{ Tfor(k)=toc; Tparfor(k)=toc; end meanTfor=mean(Tfor) meanTfor = 0.2513 meanTparfor=mean(Tparfor) % First call very slow, setup of pool with workers meanTparfor = 0.5459 % Still 2 x slower, gosh! %}
figure fplot(f,[lb(1),ub(end)]) hold on plot(xmin,ymin,'*r');grid on;shg
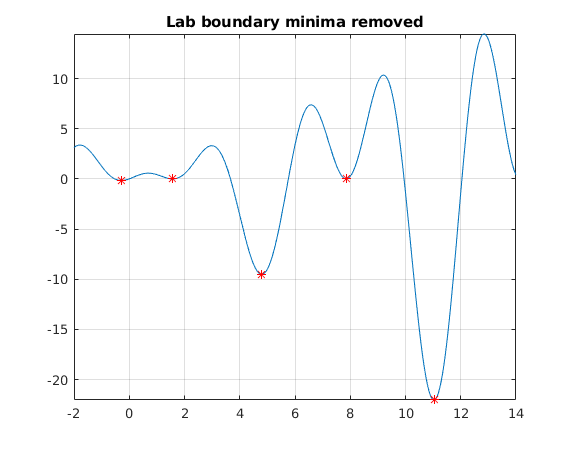
Remove boundarypoints
Here (and often in general) at least some of the boundarypoints are artficial, for the original minimzation problem. Let's remove them.
minpts([2 6 8],:)=[] figure fplot(f,[lb(1),ub(end)]) hold on title('Lab boundary minima removed') plot(minpts(:,1),minpts(:,2),'r*');grid on;shg
minpts =
-0.2809 -0.1599
1.5708 0.0000
4.7954 -9.5084
7.8540 0.0000
11.0318 -22.0274